| Immuno-Genomics and Systems Biology |
| Publications in peer-reviewed Journals / patents:
List of publications:
- Wadhwa K, Malik S, Balaji S, Thiruvengadam R, Bashyam MD, Bhattacharya PK, Behera B, Bhardwaj P, Biswas NK, Das A, Dey A, Dhotre D, Dias M, Dubey P, Dutta S, Gadepalli R, Gosain M, Goud KI, Gupta NK, Gupta N, Jana P, Jena D, John E, Karunanidhi A, Khan SMS, Khattar S, Paul APK, Kumar S, Maitra A, Modi N, Moorthy M, Nagaraj S, Palakodeti D, Pandey AK, Pandey A, Raghav SK, Ramasubban S, Raghavan S, Harikrishnan S, Krishnamoorthy S, Selvamurugan S, Sardana R, Shouche Y, Singh A, Singh AK, Ramasubramaniyan V, Yadav M, Zahoor D, Narreddy S, Bhatnagar S, Wadhwa N, Das B, Garg PK. orrelation of severity & clinical outcomes of COVID-19 with virus variants: A prospective, multicentre hospital network study. Indian J Med Res, 2024 Feb 9.
- Singh P, Sen K, Sa P, Khuntia A, Raghav SK, Swain RK, Sahoo SK. Piperlongumine based nanomedicine impairs glycolytic metabolism in triple negative breast cancer stem cells through modulation of GAPDH & FBP1. Phytomedicine, 2024 Jan:123:155181.
- Sen K, Biswas VK, Ghosh A, Prusty S, Nayak SP, Podder S, Gupta B,Raghav SK. NCoR1: a key player regulating mycobacterium tuberculosis Autophagy 2023 Nov 12:1-2.
- Biswas VK, Sen K, Ahad A, Jha A, Ghosh A, Prusty S, Swain M, Kumar D, Gupta Bhawna, Raghav SK. NCoR1 controls Mycobacterium tuberculosis growth in myeloid cells by regulating AMPK-mTOR-TFEB axis. PLoS Biology, 2023 Aug 17;21(8):e3002231.
- Tripathy A, Swain N, Padhan P, Raghav SK, Gupta B. Lactobacillus rhamnosus reduces CD8+T cell mediated inflammation in patients with rheumatoid arthritis. Immunobiology. 2023 Jul; 228: 152415.
- Mallik S, Dodia H, Ghosh A, Srinivasan R, Good L, Raghav SK, Beuria TK. FtsE, the Nucleotide Binding Domain of the ABC Transporter Homolog FtsEX, Regulates Septal PG Synthesis in E. coli. Microbiol Spectr. 2023 Apr 4:e0286322.
- Sen K, Pati R, Jha A, Mishra GP, Prusty S, Chaudhary S, Swetalika S, Podder S, Sen A, Swain M, Nanda RK, Raghav SK. NCoR1 controls immune tolerance in conventional dendritic cells by fine-tuning glycolysis and fatty acid oxidation. Redox Biol. 2023, 59:102575.
- Jha A, Ahad A, Mishra GP, Sen K, Sen A, Smita S, Minz AP, Biswas VK, Tripathy A, Senapati S, Gupta B, Acha-Orbea H, Raghav SK. SMRT and NCoR1 fine-tune inflammatory versus tolerogenic balance in dendritic cells by differentially regulating STAT3 signaling. Front Immunol. 2022, September 22.
- Mohapatra P, Mohanty S, Ansari SA, Shriwas O, Ghosh A, Rath R, Majumdar SKD, Swain RK, Raghav SK, Dash R. CMTM6 attenuates cisplatin-induced cell death in OSCC by regulating AKT/c-Myc-driven ribosome biogenesis. FASEB J. 2022 36: e22566.
- Parai D, Choudhary HR, Dash GC, Behera S, Mishra N, Pattnaik D, Raghav SK, Mishra SK, Sahoo SK, Swain A, Mohapatra I, Pattnaik M, Moharana A, Jena SR, Praharaj I, Subhadra S, Kanungo S, Bhattacharya D, Pati S. Dynamicity and persistence of severe acute respiratory syndrome coronavirus-2 antibody response after double dose and the third dose with BBV-152 and AZD1222 vaccines: A prospective, longitudinal cohort study. Front Microbiol. 2022 13: 942659.
- Mishra GP, Jha A, Ahad A, Sen K, Sen A, Podder S, Prusty S, Biswas VK, Gupta B, Raghav SK*. Epigenomics of conventional type-I dendritic cells depicted preferential control of TLR9 versus TLR3 response by NCoR1 through differential IRF3 activation. Cell Mol Life Sci. 2022, 79: 429.
- Prasad P, Mahapatra S, Mishra R, Murmu KC, Aggarwal S, Sethi M, Mohapatra P, Ghosh A, Yadav R, Dodia H, Ansari SA, De S, Singh D, Suryawanshi A, Dash R, Senapati S, Beuria TK, Chattopadhyay S, Syed GH, Swain R, Raghav SK, Parida A. Long-read 16S-seq reveals nasopharynx microbial dysbiosis and enrichment of Mycobacterium and Mycoplasma in COVID-19 patients: a potential source of co-infection. Mol Omics. 2022, 18: 490-505.
- Singh B, Avula K, Chatterjee S, Datey A, Ghosh A, De S, Keshry SS, Ghosh S, Suryawanshi AR, Dash R, Senapati S, Beuria TK, Prasad P, Raghav SK, Swain R, Parida A, Hussain Syed G*, Chattopadhyay S*. Isolation and Characterization of Five Severe Acute Respiratory Syndrome Coronavirus 2 Strains of Different Clades and Lineages Circulating in Eastern India. Front Microbiol. 2022, 13: 856913.
- Ghosh A, Walia S, Rattan R, Kanampalliwar A, Jha A, Aggarwal S, Fatma S, Das N, Chayani N, Prasad P, Raghav SK*, Parida A. Genomic profiles of vaccine breakthrough SARS-CoV-2 strains from Odisha, India. Int J Infect Dis. 2022, 119: 111-113.
- Kemp SA, Cheng MTK, Hamilton WL, Kamelian K; Indian SARS-CoV-2 Genomics Consortium (INSACOG), Singh S, Rakshit P, Agrawal A, Illingworth CJR, Gupta RK*. Transmission of B.1.617.2 Delta variant between vaccinated healthcare workers. Sci Rep. 2022, 12: 10492.
- Swain N, Tripathy A, Padhan P, Raghav SK, Gupta B. Toll-like receptor-7 activation in CD8+ T cells modulates inflammatory mediators in patients with rheumatoid arthritis. Rheumatol Int. 2022, 42: 1235-1245.
- Pathak AK, Mishra GP, Fatihi S, Abbas T, Uppili B, Ghosh A, Banu S, Bhoyar RC, Jain R, Divakar MK, Imran M, Faruq M, Sowpati DT, Thukral L, Raghav SK*, Mukerji M*. Spatio-temporal dynamics of intra-host variability in SARS-CoV-2 genomes. Nucleic Acid Research 2022, 50: 1551-1561.
- Chatterjee S, Datey A, Sengupta S, Ghosh A, Jha A, Walia S, Singh S, Suranjika S, Bhattacharya G, Laha E, Keshry SS, Ray A, Pani SS, Suryawanshi AR, Dash R, Senapati S, Beuria TK, Syed GH, Prasad P, Raghav SK, Devadas S, Swain RK, Chattopadhyay S, Parida A. Clinical, Virological, Immunological, and Genomic Characterization of Asymptomatic and Symptomatic Cases With SARS-CoV-2 Infection in India. Front Cell Infect Microbiol. 2021, 11: 725035.
- Sen K, Datta S, Ghosh A, Jha A, Ahad A, Chatterjee S, Suranjika S, Sengupta S, Bhattacharya G, Shriwas O, Avula K, Kshatri J, Prasad P, Swain R, Parida AK, Raghav SK*. Single-Cell Immunogenomic Approach Identified SARS-CoV-2 Protective Immune Signatures in Asymptomatic Direct Contacts of COVID-19 Cases. Front Immunol. 2021 12: 733539.
- Suresh V, Mohanty V, Avula K, Ghosh A, Singh B, Reddy RK, Parida D, Suryawanshi AR, Raghav SK, Chattopadhyay S, Prasad P, Swain RK, Dash R, Parida A, Syed GH, Senapati S*. Quantitative proteomics of hamster lung tissues infected with SARS-CoV-2 reveal host factors having implication in the disease pathogenesis and severity. FASEB J. 2021, 7: e21713.
- Choudhary HR, Parai D, Chandra Dash G, Kshatri JS, Mishra N, Choudhary PK, Pattnaik D, Panigrahi K, Behera S, Ranjan Sahoo N, Podder S, Mishra A, Raghav SK, Mishra SK, Pradhan SK, Sahoo SK, Pattnaik M, Rout UK, Nanda RR, Mondal N, Kanungo S, Palo SK, Bhattacharya D, Pati S. Persistence of Antibodies Against Spike Glycoprotein of SARS-CoV-2 in Healthcare Workers Post Double Dose of BBV-152 and AZD1222 Vaccines. Front Med (Lausanne). 2021, 8: 778129.
- Bhattacharya D, Choudhary HR, Parai D, Dash GC, Kshatri JS, Mishra N, Choudhary PK, Pattnaik D, Panigrahi K, Behera S, Sahoo NR, Podder S, Mishra A, Raghav SK, Mishra SK, Pradhan SK, Sahoo SK, Pattnaik M, Rout UK, Ranjan R, Mondal N, Kanungo S, Palo SK, Pati S*. Persistence of antibodies against Spike glycoprotein of SARS-CoV-2 in health care workers post double dose of BBV-152 and AZD1222 vaccines. Frontiers in Medicine 2021, 8: 778129.
- Dhar MS, Marwal R, Vs R, Ponnusamy K, Jolly B, Bhoyar RC, Sardana V, Naushin S, Rophina M, Mellan TA, Mishra S, Whittaker C, Fatihi S, Datta M, Singh P, Sharma U, Ujjainiya R, Bhatheja N, Divakar MK, Singh MK, Imran M, Senthivel V, Maurya R, Jha N, Mehta P, A V, Sharma P, Vr A, Chaudhary U, Soni N, Thukral L, Flaxman S, Bhatt S, Pandey R, Dash D, Faruq M, Lall H, Gogia H, Madan P, Kulkarni S, Chauhan H, Sengupta S, Kabra S; Indian SARS-CoV-2 Genomics Consortium (INSACOG)‡, Gupta RK, Singh SK, Agrawal A, Rakshit P, Nandicoori V, Tallapaka KB, Sowpati DT, Thangaraj K, Bashyam MD, Dalal A, Sivasubbu S, Scaria V, Parida A, Raghav SK, Prasad P, Sarin A, Mayor S, Ramakrishnan U, Palakodeti D, Seshasayee ASN, Bhat M, Shouche Y, Pillai A, Dikid T, Das S, Maitra A, Chinnaswamy S, Biswas NK, Desai AS, Pattabiraman C, Manjunatha MV, Mani RS, Arunachal Udupi G, Abraham P, Atul PV, Cherian SS*. Genomic characterization and epidemiology of an emerging SARS-CoV-2 variant in Delhi, India. 2021, 374: 995-999.
- Mlcochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, Datir R, Collier DA, Albecka A, Singh S, Pandey R, Brown J, Zhou J, Goonawardane N, Mishra S, Whittaker C, Mellan T, Marwal R, Datta M, Sengupta S, Ponnusamy K, Radhakrishnan VS, Abdullahi A, Charles O, Chattopadhyay P, Devi P, Caputo D, Peacock T, Wattal C, Goel N, Satwik A, Vaishya R, Agarwal M; Indian SARS-CoV-2 Genomics Consortium (INSACOG); Genotype to Phenotype Japan (G2P-Japan) Consortium; CITIID-NIHR BioResource COVID-19 Collaboration, Mavousian A, Lee JH, Bassi J, Silacci-Fegni C, Saliba C, Pinto D, Irie T, Yoshida I, Hamilton WL, Sato K, Bhatt S, Flaxman S, James LC, Corti D, Piccoli L, Barclay WS, Rakshit P, Agrawal A, Gupta RK*. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. 2021, 599: 114-119.
- Borah NA, Sradhanjali S, Barik MR, Jha A, Tripathy D, Kaliki S, Rath S, Raghav SK, Patnaik S, Mittal R, Reddy MM. Aurora Kinase B Expression, Its Regulation and Therapeutic Targeting in Human Retinoblastoma. Invest Ophthalmol Vis Sci. 2021, 62: 16.
- Tripathy A, Padhan P, Swain N, Raghav SK, Gupta B. Increased Extracellular ATP in Plasma of Rheumatoid Arthritis Patients Activates CD8+T Cells. Arch Med Res. 2021, S0188-4409(21)00001-1.
- Dash P, Turuk J, Behera SK, Palo SK, Raghav SK*, Ghosh A, Sabat J, Rath S, Subhadra S, Rana K, Bhattacharya D, Kanungo S, Kshatri JS, Mishra BK, Dash S, Parida A, Pati S*. Sequence analysis of Indian SARS-CoV-2 isolates shows a stronger interaction of mutant receptor-binding domain with ACE2. Int J Infect Dis. 2021, 104: 491-500.
- Mohapatra P, Shriwas O, Mohanty S, Ghosh A, Smita S, Kaushik SR, Arya R, Rath R, Das Majumdar SK, Muduly DK, Raghav SK, Nanda RK, Dash R. CMTM6 drives cisplatin resistance by regulating Wnt signaling through the ENO-1/AKT/GSK3β axis. JCI Insight. 2021, 6: e143643.
- Smita S, Ghosh A, Biswas VK, Ahad A, Podder S, Jha A, Sen K, Acha-Orbea H, Raghav SK*. Zbtb10 transcription factor is crucial for murine cDC1 activation and cytokine secretion. Eur J Immunol. 2021 Feb 1.
- Raghav S*, Ghosh A, Turuk J, Kumar S, Jha A, Madhulika S, Priyadarshini M, Biswas VK, Shyamli PS, Singh B, Singh N, Singh D, Datey A, Avula K, Smita S, Sabat J, Bhattacharya D, Kshatri JS, Vasudevan D, Suryawanshi A, Dash R, Senapati S, Beuria TK, Swain R, Chattopadhyay S, Syed GH, Dixit A, Prasad P; Odisha COVID-19 Study Group; ILS COVID-19 Team, Pati S, Parida A. Analysis of Indian SARS-CoV-2 Genomes Reveals Prevalence of D614G Mutation in Spike Protein Predicting an Increase in Interaction With TMPRSS2 and Virus Infectivity. Front Microbiol. 2020, 11: 594928.
- Jaiswal KS, Khanna S, Ghosh A, Padhan P, Raghav SK, Gupta B. Differential mitochondrial genome in patients with Rheumatoid Arthritis. 2021, 54: 1-12.
- Ahad A, Smita S, Mishra GP, Biswas VK, Sen K, Gupta B, Garcin D, Acha-Orbea H, Raghav SK. NCoR1 fine-tunes type-I IFN response in cDC1 dendritic cells by directly regulating Myd88-IRF7 axis under TLR9. Eur J Immunol. 2020, 50: 1959-1975.
- Khanna S, Pradhan P, Jaiswal KS, Jain AP, Ghosh A, Tripathy A, Gowda H, Raghav SK, Gupta B. Altered mitochondrial proteome and functional dynamics in patients with rheumatoid arthritis. Mitochondrion 2020, 54:8-14.
- Das D, Das A, Sahu M, Mishra SS, Khan S, Bejugam PR, Rout PK, Das A, Bano S, Mishra GP, Raghav SK, Dixit A, Panda AC. Identification and characterization of circular intronic RNAs derived from insulin gene. International Journal of Molecular Sciences 2020, 21: 4302.
- Mishra GP, Ghosh A, Jha A, Raghav SK. BedSect: An integrated web server application to perform intersection, visualization and functional annotation of genomic regions from multiple datasets. Frontiers in Genetics 2020, 11: 3.
- Chen W, Schwalie PC, Pankevich EV, Gubelmann C, Raghav SK, Dainese R, Cassano M, Imbeault M, Jang SM, Russeil J, Delessa T, Duc J, Trono D, Wolfrum C, Deplancke B*. ZFP30 promotes adipogenesis through the KAP1-mediated activation of a retrotransposon-derived Pparg2 enhancer. Nat Commun. 2019, 10: 1809.
- Ahad A, Stevanin M, Smita S, Mishra GP, Gupta D, Waszak S, Sarkar UA, Basak S, Gupta B, Acha-Orbea H, Raghav SK*. NCoR1: putting the brakes on the dendritic cell immune tolerance. iScience. 2019, 996-1011
- Smita S, Ahad A, Ghosh A, Biswas VK, Koga MM, Gupta B, Acha-Orbea H, Raghav SK. Importance of EMT Factor ZEB1 in cDC1 “MutuDC Line” Mediated Induction of Th1 Immune Response. Frontiers in Immunology 2018, 9: 2604.
- Jena KK, Kolapalli SP, Mehto S, Nath P, Das B, Sahoo PK, Ahad A, Syed GH, Raghav SK, Senapati S, Chauhan S, Chauhan S. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018, 37: e98358.
- Rib L, Villeneuve D, Minocha S, Praz V, Hernandez N, Guex N, Herr W; CycliX Consortium. Cycles of gene expression and genome response during mammalian tissue regeneration. Epigenetics Chromatin.2018, 11: 52.
- Tripathy A, Khanna S, Padhan P, Smita S, Raghav SK, Gupta B. Direct recognition of LPS drive TLR4 expressing CD8+ T cell activation in patients with rheumatoid arthritis. Sci Rep. 2017, 7: 933.
- Jonathan Aryeh Sobel, Irina Krier, Teemu Andersin, Sunil Raghav, Donatella Canella, Federica Gilardi, Alexandra Styliani Kalantzi, Guillaume Rey, Benjamin Weger, Frédéric Gachon, Matteo Dal Peraro, Nouria Hernandez, Ueli Schibler, Bart Deplancke, Felix Naef, and the CycliX consortium. Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biology 2017, 15: e2001069.
- Rachana Pradhan, Johannes Bues, Vincent Gardeux, Petra C. Schwalie, Daniel Alpern, Wanze Chen, Julie Russeil, Sunil Raghav, and Bart Deplancke. Dissecting the brown adipogenic regulatory network using integrative genomics. Scientific Reports 2017, 7:42130.
- Raghav SK, Gupta B*. Single cell Transcriptomics for autoimmune disorders. J Mol Genet Med 2015, 9: 191.
- Sahu D, Raghav SK, Gautam H, Das HR. A novel coumarin derivative, 8-methoxy chromen-2-one alleviates collagen induced arthritis by down regulating nitric oxide, NFκB and proinflammatory cytokines. Immunopharmacol. 2015, 29, 891-900.
- Jain S, Suklabaidya S, Das B, Raghav SK, Batra SK, Senapati S. TLR4 activation by lipopolysaccharide confers survival advantage to growth factor deprived prostate cancer cells. 2015, 75:1020-1033.
- Waszak SM, Delaneau O, Gschwind AR, Kilpinen H, Raghav SK, Witwicki RM, Orioli A, Wiederkehr M, Panousis NI, Yurovsky A, Romano-Palumbo L, Planchon A, Bielser D, Padioleau I, Udin G, Thurnheer S, Hacker D, Hernandez N, Reymond A, Deplancke B, Dermitzakis ET. Population Variation and Genetic Control of Modular Chromatin Architecture in Humans. Cell 2015, 27: 1039-50.
- Gilardi F, Migliavacca E, Naldi A, Baruchet M, Canella D, Le Martelot G, Guex N, Desvergne B; CycliX Consortium. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014, 10: e1004155.
- Gubelmann C, Schwalie PC, Raghav SK, Röder E, Delessa T, Kiehlmann E, Waszak SM, Corsinotti A, Udin G, Holcombe W, Rudofsky G, Trono D, Wolfrum C, Deplancke B. Identification of the transcription factor ZEB1 as a central component of the adipogenic gene regulatory network. 2014: e03346.
- Waszak SM, Kilpinen H, Gschwind AR, Orioli A, Raghav SK, Witwicki RM, Migliavacca E, Yurovsky A, Lappalainen T, Hernandez N, Reymond A, Dermitzakis ET, Deplancke B. Identification and removal of low-complexity sites in allele-specific analysis of ChIP-seq data. 2014, 30: 165-171.
- Kilpinen H, Waszak SM, Gschwind A, Raghav SK, Witwicki RM, Orioli A, Migliavacca E, Wiederkehr M, Gutierrez-Arcelus M, Panousis N, Yurovsky A, Lappalainen Tuuli, Romano-Palumbo L, Planchon A, Bielser D, Bryois J, Padioleau I, Udin G, Thurnheer S, Hacker D, Core LJ, Lis JT, Hernandez N, Reymond A, Deplancke B, Dermitzakis ET. Coordinated effects of sequence variation on DNA binding, chromatin structure, and transcription. Science 2013, 342: 744-747.
- Gubelmann C, Waszak SM, Isakova A, Holcombe W, Hens K, Iagovitina A, Feuz JD, Raghav SK, Simicevic J, Deplancke B. A yeast one-hybrid and microfluidics-based pipeline to map mammalian gene regulatory networks. Molecular Systems Biology 2013, 6; 9:682.
- Hawkins RD*, Larjo A, Tripathi SK, Wagner U, Luu Y, Loneberg T, Raghav SK, Lee LK, Lund R, Lähdesmäki H*, Ren B*, Lahesmaa R* Global chromatin state analysis reveals lineage-specific enhancers during early cell fate specification of human Th1 and Th2 cells. Immunity 2013, 38:1271-1284.
- Simicevic J, Schmid AW, Gilardoni P, Zoller B, Raghav SK, Krier I, Gubelmann C, Lisacek F, Naef F, Moniatte M, Lisacek F, Deplancke B. Absolute copy number analysis of transcription factors during cellular differentiation using a multiplex, targeted proteomics approach. Nature Methods 2013, 10: 570-576.
- LeMartelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, Herr W, Deplancke B, Schibler U, Rougemont J, Guex N, Hernandez N*, Naef F*, and CycliX consortium including Raghav SK. Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during circadian cycles. PLoS Biology 2012, 10: e1001442.
- Raghav SK, Waszak MS, Krier I, Gubelmann C, Mikkelsen ST, Deplancke B*. Integrative genomics identifies SMRT a gatekeeper of early adipogenesis through transcription factors C/EBPβ and KAISO. Molecular Cell 2012, 46, 335–350.
- Bernasconi D, Canella D, Gilardi F, LeMartelot GL, Praz V, Migliavacca E, Delorenzi M, Hernandez N*, and CycliX consortium including Raghav SK. A Multiplicity of Factors Contributes to Selective Transcription of a Subset of RNA polymerase III Genes in Mouse Liver. Genome Research 2012, 22: 666-680.
- Baitsch L, Baumgaertner P, Devêvre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, Rufer N, Speiser DE*. Exhaustion of human self/tumor-specific CD8 T-cells in melanoma metastases. Journal of Clinical Investigation 2011, 121: 2350-60.
- Elo LL, Järvenpää H, Tuomela S, Raghav SK, Ahlfors H, Laurila K, Gupta B, Lund RJ, Tahvanainen J, Hawkins RD, Orešič M, Lähdesmäki H, Rasool O, Rao KV, Aittokallio T, Lahesmaa R*. IL-4 and STAT6-mediated transcriptional regulation to initiate Th2 program in human T cells. Immunity 2010, 6: 852-862.
- Laajala TD, Raghav S, Tuomela S, Lahesmaa R, Aittokallio T, Elo LL*. A practical comparison of methods for detecting transcription factor binding sites in ChIP-seq experiments. BMC Genomics, 2009, 18: 618.
- Rautajoki KJ, Kyläniemi MK, Raghav SK, Rao K, Lahesmaa R*. An insight into molecular mechanisms of human T helper cell differentiation. Annals of Medicine 2008, 40: 322-35.
- Gupta B, Raghav SK, Das HR*. S-Nitrosylation of mannose binding lectin regulates its functional activities and the formation of autoantibody in rheumatoid arthritis. Nitric oxide: Biology and Chemistry 2008, 18: 266-273.
- Raghav SK, Gupta B, Das HR*. Inhibition of lipopolysaccharide inducible iNOS and IL-1b expression by active compounds isolated from Ruta graveolens L. through suppression of NF-kB activation in murine macrophages. European Journal of Pharmacology 2007, 560: 69-80.
- Raghav SK, Gupta B, Agrawal C, Chaturvedi VP, Das HR*. Expression of TNF-a and related signaling molecules in the peripheral blood mononuclear cells of rheumatoid arthritis patients. Mediators of Inflammation 2006, 12682: 1-6.
- Raghav SK, Gupta B, Agrawal C, Saroha A, Das RH, Chaturvedi VP, Das HR*. Altered expression and glycosylation of plasma proteins in rheumatoid arthritis. Glycoconjugate Journal 2006, 23: 169-175.
- Raghav SK, Gupta B, Agrawal C, Goswami K, Das HR*. Anti-inflammatory effect of Ruta graveolens L. in murine macrophage cells. Journal of Ethnopharmacology 2006, 104: 234-239.
- Gupta B, Raghav SK, Agrawal C, Das RH, Chaturvedi VP, Das HR*. Anti-MBL autoantibodies in patients with rheumatoid arthritis: prevalence and clinical significance. Journal of Autoimmunity 2006, 27: 125-133.
- Chaudhury I, Raghav SK, Gautam H, Das HR, Das RH*. Suppression of inducible nitric oxide synthase by 10-23 DNAzymes in murine macrophages. FEBS Letters 2006, 580: 2046-2052.
- Gupta B, Agrawal C, Raghav SK, Das SK, Das RH, Chaturvedi VP, Das HR*. Association of mannose-binding lectin gene (MBL2) polymorphisms with rheumatoid arthritis in an Indian cohort of case-control samples. Journal of Human Genetics 2005, 50: 583–591.
- Agrawal C, Raghav SK, Gupta B, Das RH, Chaturvedi VP, Goswami K, Das HR*. Tumor necrosis factor-alpha microsatellite polymorphism association with rheumatoid arthritis in Indian patients. Archives of Medical Research 2005, 36: 555-559.
-
- Expanding research horizons abroad, annual magazine of Turku University, 2008.
- Genetic variability of SARS-CoV-2 strains and its impact on virus transmission, Atimukta Jha & Sunil K. Raghav, 2021.Article in University magazine / Science Horizon magazine
-
Book Chapters / Article in University magazine / Science Horizon magazine
- Das HR*, Raghav S, Gupta B and Das RH. Anti-inflammatory compounds from medicinal plant Ruta graveolens. Acta Horticulture 2007, 756: 389- 398.
- Raghav SK, Deplancke B*. Genome-wide profiling of DNA binding proteins using barcode-based multiplex Solexa sequencing. Methods in Molecular Biology 2012, 786: 247-62.
- Expanding research horizons abroad, annual magazine of Turku University, 2008.
- Genetic variability of SARS-CoV-2 strains and its impact on virus transmission, Atimukta Jha & Sunil K. Raghav, 2021.
-
Patents Filed / granted:
- Method for the diagnosis of rheumatoid arthritis. Hasi Das, Bhawna Gupta, Sunil Kumar Raghav, Kalyan Goswami, Charu Agrawal and Rakha Hari Das. IPC8 Class: AG01N3353FI, US Patent 7,763,436.
- Novel anti-inflammatory compound 3-(1,1-dimethyl-allyl)-6-hydroxy-chromen-2-one isolated from Ruta graveolens L. Hasi Das, Sunil Kumar Raghav, Bhawna Gupta. IPC8 Class: AA61K31352FI, US Patent App. 11/935,985.
- Novel diagnostic marker, a diagnostic kit and a method for diagnosis of rheumatoid arthritis. Hasi Das, Bhawna Gupta, Sunil Kumar Raghav, Kalyan Goswami, Charu Agrawal and Rakha Hari Das. US Patent App. 11/281,243.
|
|
|


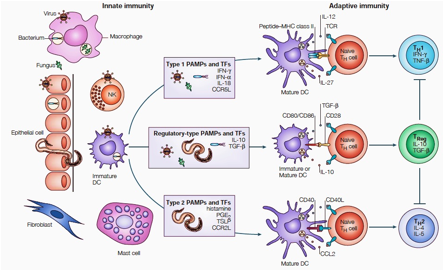 response generation in DCs, (ii) Engineer DCs to immune-modulate DC immune responses to fine-tune T cell speciation/differentiation, and (iii) Extrapolate the potential of engineered DCs for immune-therapeutic approaches.
response generation in DCs, (ii) Engineer DCs to immune-modulate DC immune responses to fine-tune T cell speciation/differentiation, and (iii) Extrapolate the potential of engineered DCs for immune-therapeutic approaches.