2020
- Kolapalli SP, Sahu R, Chauhan NR, Jena KK, Mehto S, Das SK, Jain A, Rout M, Dash R, Swain RK, Lee DY, Rusten TE, Chauhan S*, Chauhan S*. RNA binding RING E3-ligase DZIP3/hRUL138 is a novel driver of cell cycle and cancer progression by employing a unique mechanism to stabilize Cyclin D1. Cancer Research, 2020. PMID: 33067265. * Corresponding author
- Jena KK, Mehto S, Nath P, Chauhan NR, Sahu R, Dhar K, Das SK, Kolapalli SP, Murmu KC, Jain A, Krishna S, Sahoo BS, Chattopadhyay S, Rusten TE, Prasad P, Chauhan S, Chauhan S*. Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. EMBO Rep. 2020 Jul 27:e50051. doi: 10.15252/embr.202050051.* Corresponding author
- Nath P, Jena KK, Mehto S, Chauhan NR, Sahu R, Dhar K, Srinivas K, Chauhan S, Chauhan S*. IRGM Links Autoimmunity to Autophagy. Autophagy. 2020 Aug 19. doi: 10.1080/15548627.2020.1810920. PMID: 32813580. Corresponding author
2019
- Mehto S, Jena KK, Nath P, Chauhan S, Kolapalli SP, Das SK, Sahoo PK, Jain A, Taylor GA, Chauhan S*. The Crohn’s disease risk factor IRGM limits NLRP3 inflammasome activation by impeding its assembly and by mediating its selective autophagy. Molecular Cell. 2019 Feb 7;73(3):429-445.e7. Preview: Nabar NR, Kehrl JH. Inflammasome Inhibition Links IRGM to Innate Immunity. Mol Cell. 2019 Feb 7;73(3):391-392.
- Mehto S, Chauhan S, Jena KK, Chauhan NR, Nath P, Sahu R, Dhar K, Das SK, Chauhan S*. IRGM restrains NLRP3 inflammasome activation by mediating its SQSTM1/p62-dependent selective autophagy. Autophagy. 2019 Jun 20:1-3.
- Jena KK, Mehto S, Kolapalli SP, Nath P, Sahu R, Chauhan NR, Sahoo PK, Dhar K, Das SK, Chauhan S, Chauhan S*. TRIM16 governs the biogenesis and disposal of stress-induced protein aggregates to evade cytotoxicity: implication for neurodegeneration and cancer. Autophagy. 2019 May;15(5):924-926.
- Jena KK, Mehto S, Kolapalli SP, Nath P, Chauhan S, Chauhan S*. TRIM16 employs NRF2, ubiquitin system and aggrephagy for safe disposal of stress-induced misfolded proteins. Cell Stress. 2018 Nov 16;2(12):365-367.
- Saha S, Murmu KC, Biswas M, Chakraborty S, Basu J, Madhulika S, Kolapalli SP, Chauhan S, Sengupta A, Prasad P. Transcriptomic Analysis Identifies RNA Binding Proteins as Putative Regulators of Myelopoiesis and Leukemia. Front Oncol. 2019 Aug 6;9:692.
2018
- Jena KK, Kolapalli SP, Mehto S, Nath P, Das B, Sahoo PK, Ahad A, Syed GH, Raghav SK, Senapati S, Chauhan S, Chauhan S. TRIM16 controls assembly and degradation of protein aggregates by modulating the p62-NRF2 axis and autophagy. EMBO J. 2018 Sep 14;37(18).
- Jena KK, Kolapalli SP, Mehto S, Chauhan S, Chauhan S. TRIM16 controls turnover of protein aggregates by modulating NRF2, ubiquitin system, and autophagy: implication for tumorigenesis. 22 Oct 2018, Molecular & Cellular Oncology. 2018 Oct 22;5(6):e1532251
- Jena KK, Mehto S, Kolapalli SP, Nath P, Sahu R, Chauhan NR, Sahoo PK, Dhar K, Das SK, Chauhan S, Chauhan S. TRIM16 governs the biogenesis and disposal of stress-induced proteins aggregates to evade cytotoxicity: implication in neurodegeneration and cancer. Autophagy. 2019 Feb 26:1-3.
- Jena KK, Mehto S, Kolapalli SP, Nath P, Chauhan S, Chauhan S. TRIM16 employs NRF2, ubiquitin system and aggrephagy for safe disposal of stress-induced misfolded protein, Cell Stress, 2, No. 12, pp. 365 – 367.
2017
- Kumar S, Chauhan S, Jain A, Ponpuak M, Choi SW, Mudd M, Peters R, Mandell MA, Johansen T, Deretic V. Galectins and TRIMs directly interact and orchestrate autophagic response to endomembrane damage. Autophagy. 2017 Jun 3;13(6):1086-1087. doi: 10.1080/15548627.2017.1307487. Epub 2017 Apr 3. 1.
2016
- Santosh Chauhan*, Suresh Kumar*, Ashish Jain*, Marisa Ponpuak*, Michal H. Mudd, Tomonori Kimura,Seong Won Choi, Ryan Peters, Michael Mandell, Terje Johansen, and Vojo Deretic. TRIMs and Galectins globally cooperate and TRIM16 and Galectin-3 codirect autophagy in endomembrane damage homeostasis. * Equal authorships. Developmental Cell.
- Klionsky DJ et al.,. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy Jan 2;12(1):1-222, 2016
2015
- Santosh Chauhan*, Zahra Ahmed*, Steven B. Bradfute,….Vincent Piguet, and Vojo Deretic. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. * Equal authorship. Nature Communication. 2015 Oct 27;6:8620. doi: 10.1038/ncomms9620.
- Chauhan, S.*, Mandell, M. and Deretic, V.*. IRGM governs the core autophagy machinery to conduct antimicrobial defense. Volume 58, Issue 3, p507–521, 7 May, Molecular Cell. * Corresponding author.
- Chauhan S, Mandell MA, Deretic V. Mechanism of action of the tuberculosis and Crohn disease risk factor IRGM in autophagy. Autophagy. 2015 Aug 27:0. PMID: 26313894.
- Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015 May 16;35:106-116. doi: 10.1016/j.ceb.2015.04.016.
- Deretic V, Kimura T, Timmins G, Moseley P, Chauhan S, Mandell M. Immunological manifestations of autophagy. J Clin Invest. 2015 Jan;125(1):75-84. doi: 10.1172/JCI73945.
2014
- Mandell MA, Jain A, Arko-Mensah J, Chauhan S, Kimura T, Dinkins C, Silvestri G, Münch J, Kirchhoff F, Simonsen A, Wei Y, Levine B, Johansen T, Deretic V. TRIM proteins regulate autophagy and can target autophagic substrates by direct recognition. Developmental Cell. 2014 Aug 25; 30(4):394-409.
- Dupont N, Chauhan S, Arko-Mensah J, Castillo EF, Masedunskas A, Weigert R, Robenek H, Proikas-Cezanne T, Deretic V. Neutral lipid stores and lipase PNPLA5 contribute to autophagosome biogenesis. Current Biology. 2014 Mar 17;24 (6):609-20.
2013
- Chauhan, S., Goodwin, J.G., Chauhan, S., Manyam, G., Wang, J., Kamat, A.M., and Boyd, D.D. (2013). ZKSCAN3 Is a Master Transcriptional Repressor of Autophagy. Molecular Cell. Apr 11;50(1):16-28. Highlighted in Cancer Discovery (AACR) journal, February 28, 2013; doi:10.1158/2159-8290.CD-RW2013-046. Recommended by Faculty of 1000 (http://f1000.com/prime/717980675)
- Bradfute SB, Castillo EF, Arko-Mensah J, Chauhan S, Jiang S, Mandell M, Deretic V. Autophagy as an immune effector against tuberculosis. Curr Opin Microbiol. 2013 Jun 18. pii: S1369-5274(13)00070-2. doi: 10.1016/j.mib.2013.05.003.
2012
- Chauhan, S., and Boyd DD. Regulation of u-PAR gene expression by H2A.Z is modulated by the MEK-ERK/AP-1 pathway. Nucleic Acids Research. 2012 Jan; 40 (2):600-13.
- Chauhan, S, Sharma, D., Singh, A., Surolia, A. and Tyagi, J.S. Comprehensive insights into Mycobacterium tuberculosis DevR (DosR) regulon activation switch. Nucleic Acids Research. 2011 Sep 1;39(17):7400-14.
2011
- Chauhan, S and Tyagi, J.S. (2011) Analysis of transcription at the oriC locus in Mycobacterium tuberculosis. Microbiol Res. 2011 Sep 20;166(6):508-14.
- Avila H, Wang H, Chauhan S, Hartig S, Boyd DD. Accelerated urokinase-receptor protein turnover triggered by interference with the addition of the glycolipid anchor. Biochem J. 2011 Mar 1;434(2):233-42.
- Gupta, R.K., Chauhan, S. and Tyagi, J.S. K182G substitution in DevR or C(8) G mutation in the Dev box impairs protein-DNA interaction and abrogates DevR-mediated gene induction in Mycobacterium tuberculosis. FEBS J. 2011 Jun; 278(12):2131-9.
- Gautam, U.S., Chauhan, S. and Tyagi, J.S. Determinants outside the DevR C-terminal domain are essential for cooperativity and robust activation of dormancy genes in Mycobacterium tuberculosis. PLoS One. 2011 Jan 27;6(1):e16500.
2010
- Majumdar, S.D., Sharma, D., Vashist, A., Kaur, K., Taneja, N.K., Chauhan, S., Challu, V.K., Ramanathan, V.D., Balasangameshwara, V., Kumar, P. et al. Co-expression of DevR and DevR(N)-Aph proteins is associated with hypoxic adaptation defect and virulence attenuation of Mycobacterium tuberculosis. PLoS One. 2010, Feb 26;5(2):e9448.
- Chauhan S, Singh A, Tyagi JS. A single-nucleotide mutation in the -10 promoter region inactivates the narK2X promoter in Mycobacterium bovis and Mycobacterium bovis BCG and has an application in diagnosis. FEMS Microbiol Lett. 2010, Feb;303(2):190-6.
2009
- Chauhan S, Tyagi JS. Powerful induction of divergent tgs1-Rv3131 genes in Mycobacterium tuberculosis is mediated by DevR interaction with a high-affinity site and an adjacent cryptic low-affinity site. Journal of Bacteriology. 2009, Oct;191(19):6075-81.
- Chauhan S, Kumar A, Singhal A, Tyagi JS, Krishna Prasad H. CmtR, a cadmium-sensing ArsR-SmtB repressor, cooperatively interacts with multiple operator sites to autorepress its transcription in Mycobacterium tuberculosis. FEBS J. 2009, Jul; 276(13):3428-39.
2008
- Chauhan S, Tyagi JS. Interaction of DevR with Multiple Binding Sites Synergistically Activates Divergent Transcription of narK2-Rv1738 Genes in Mycobacterium tuberculosis. Journal of Bacteriology. Aug 2008, p. 5394–5403, 190, No. 15.
- Chauhan S, Tyagi JS. Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: Implication in the induction of DevR target genes. Journal of Bacteriology, June 2008, p. 4301–4312, 190, No.12.
2005
- Bagchi GY, Chauhan SY, Sharma D, Tyagi JS. Transcription and autoregulation of Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology (2005), 151, 4045-4053.Y Equal contribution to work.
|
 Deretic. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. * Equal authorships. (Nat Commun. 2015 Oct 27;6:8620. doi: 10.1038/ncomms9620.)
Deretic. Pharmaceutical screen identifies novel target processes for activation of autophagy with a broad translational potential. * Equal authorships. (Nat Commun. 2015 Oct 27;6:8620. doi: 10.1038/ncomms9620.)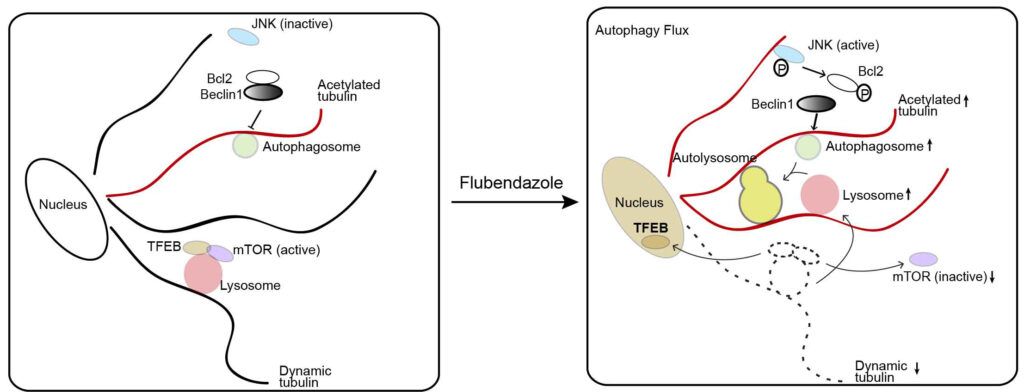 drugs) to discover new mechanisms for activation of autophagy. We identify a subset of pharmaceuticals inducing autophagic flux with effects in diverse cellular systems modelling specific stages of several human diseases such as HIV transmission and hyperphosphorylated tau accumulation in Alzheimer’s disease. One drug, flubendazole, is a potent inducer of autophagy initiation and flux by affecting acetylated and dynamic microtubules in a reciprocal way. Disruption of dynamic microtubules by flubendazole results in mTOR deactivation and dissociation from lysosomes leading to Transcription Factor EB nuclear translocation and activation of autophagy. By inducing microtubule acetylation, flubendazole activates JNK1 leading to Bcl-2 phosphorylation, causing release of Beclin-1 from Bcl-2-Beclin-1 complexes for autophagy induction, thus uncovering a new approach to inducing autophagic flux that may be applicable in disease treatment.
drugs) to discover new mechanisms for activation of autophagy. We identify a subset of pharmaceuticals inducing autophagic flux with effects in diverse cellular systems modelling specific stages of several human diseases such as HIV transmission and hyperphosphorylated tau accumulation in Alzheimer’s disease. One drug, flubendazole, is a potent inducer of autophagy initiation and flux by affecting acetylated and dynamic microtubules in a reciprocal way. Disruption of dynamic microtubules by flubendazole results in mTOR deactivation and dissociation from lysosomes leading to Transcription Factor EB nuclear translocation and activation of autophagy. By inducing microtubule acetylation, flubendazole activates JNK1 leading to Bcl-2 phosphorylation, causing release of Beclin-1 from Bcl-2-Beclin-1 complexes for autophagy induction, thus uncovering a new approach to inducing autophagic flux that may be applicable in disease treatment.

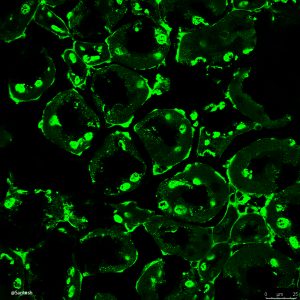
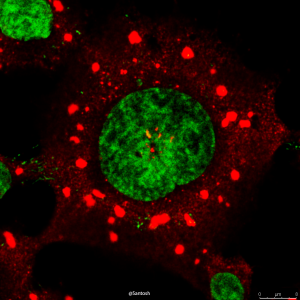
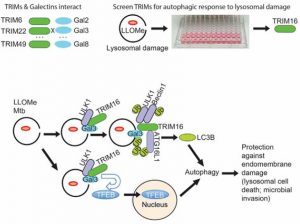 Selective autophagy performs an array of tasks to maintain intracellular homeostasis, sterility, and organellar and cellular functionality. The fidelity of these processes depends on precise target recognition and limited activation of the autophagy apparatus in a localized fashion. Here we describe cooperation in such processes between the TRIM family and Galectin family of proteins. TRIMs, which are E3 ubiquitin ligases, displayed propensity to associate with Galectins. One specific TRIM, TRIM16, interacted with Galectin-3 in a ULK1-dependent manner. TRIM16, through integration of Galectin- and ubiquitin-based processes, coordinated recognition of membrane damage with mobilization of the core autophagy regulators ATG16L1, ULK1, and Beclin 1 in response to damaged endomembranes. TRIM16 affected mTOR, interacted with TFEB, and influenced TFEB’s nuclear translocation. The cooperation between TRIM16 and Galectin-3 in targeting and activation of selective autophagy protects cells from lysosomal damage and
Selective autophagy performs an array of tasks to maintain intracellular homeostasis, sterility, and organellar and cellular functionality. The fidelity of these processes depends on precise target recognition and limited activation of the autophagy apparatus in a localized fashion. Here we describe cooperation in such processes between the TRIM family and Galectin family of proteins. TRIMs, which are E3 ubiquitin ligases, displayed propensity to associate with Galectins. One specific TRIM, TRIM16, interacted with Galectin-3 in a ULK1-dependent manner. TRIM16, through integration of Galectin- and ubiquitin-based processes, coordinated recognition of membrane damage with mobilization of the core autophagy regulators ATG16L1, ULK1, and Beclin 1 in response to damaged endomembranes. TRIM16 affected mTOR, interacted with TFEB, and influenced TFEB’s nuclear translocation. The cooperation between TRIM16 and Galectin-3 in targeting and activation of selective autophagy protects cells from lysosomal damage and
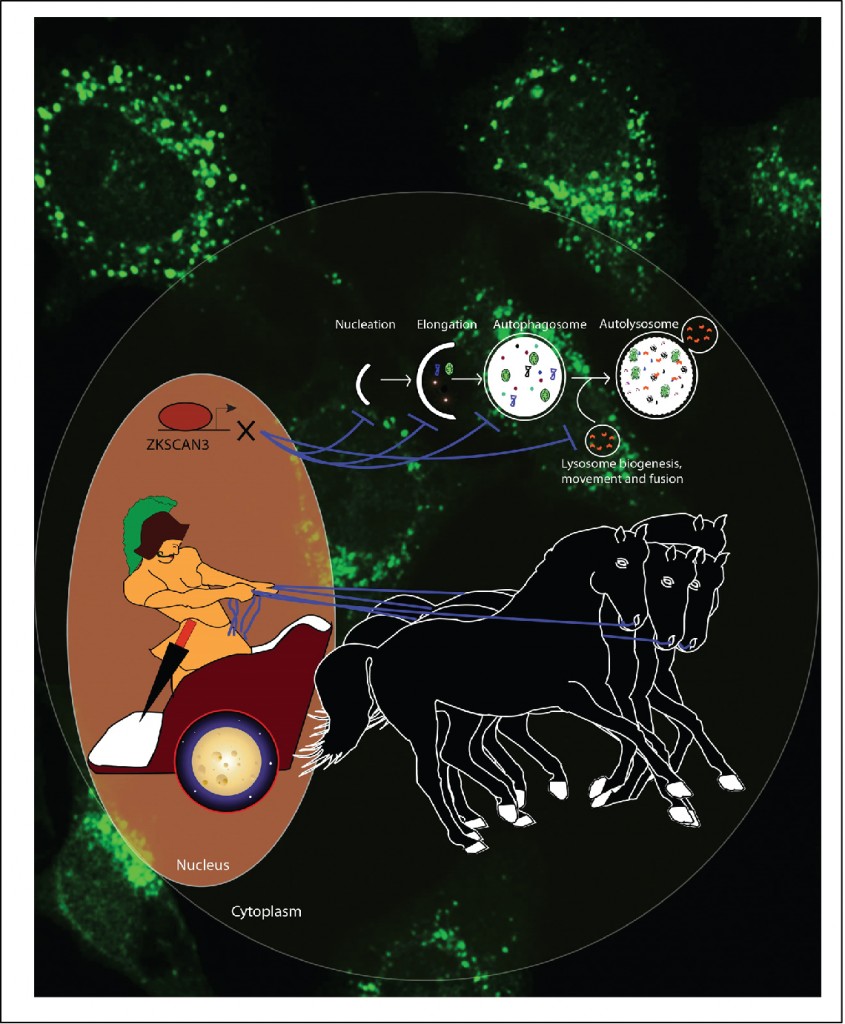 paper, we establish ZKSCAN3, a zinc finger family DNA-binding protein, as a transcriptional repressor of autophagy. Silencing of ZKSCAN3 induced autophagy and increased lysosome biogenesis. Importantly, we show that ZKSCAN3 represses transcription of a large gene set (>60) integral to, or regulatory for, autophagy and lysosome biogenesis/function and that a subset of these genes, including Map1lC3b and Wipi2, represent direct targets. Interestingly, ZKSCAN3 and TFEB are oppositely regulated by starvation and in turn oppositely regulate lysosomal biogenesis and autophagy, suggesting that they act in conjunction. Altogether, our study uncovers an autophagy master switch regulating the expression of a transcriptional network of genes integral to autophagy and lysosome biogenesis/function.
paper, we establish ZKSCAN3, a zinc finger family DNA-binding protein, as a transcriptional repressor of autophagy. Silencing of ZKSCAN3 induced autophagy and increased lysosome biogenesis. Importantly, we show that ZKSCAN3 represses transcription of a large gene set (>60) integral to, or regulatory for, autophagy and lysosome biogenesis/function and that a subset of these genes, including Map1lC3b and Wipi2, represent direct targets. Interestingly, ZKSCAN3 and TFEB are oppositely regulated by starvation and in turn oppositely regulate lysosomal biogenesis and autophagy, suggesting that they act in conjunction. Altogether, our study uncovers an autophagy master switch regulating the expression of a transcriptional network of genes integral to autophagy and lysosome biogenesis/function. This paper was Highlighted in Cancer Discovery (AACR) journal, February 28,
This paper was Highlighted in Cancer Discovery (AACR) journal, February 28,