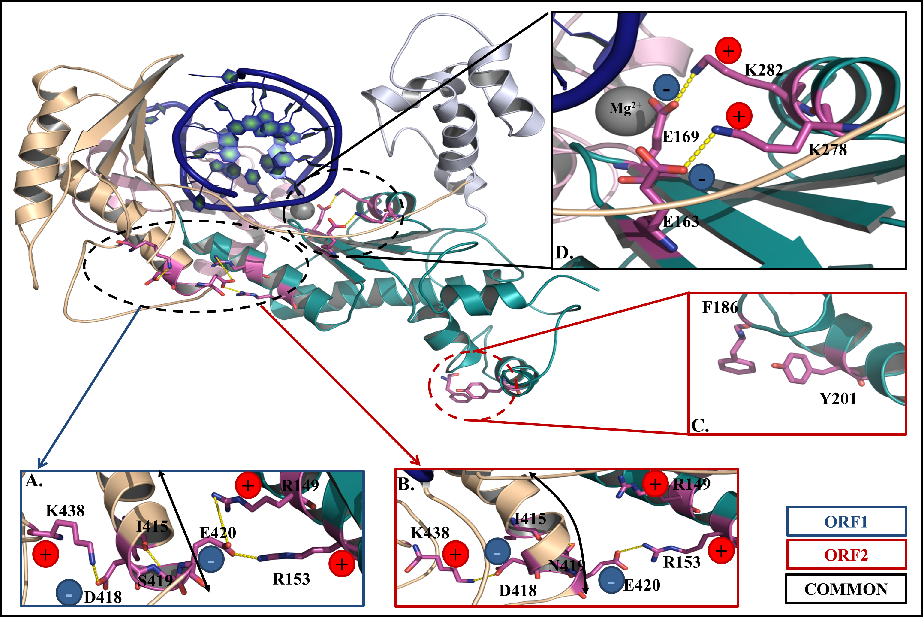
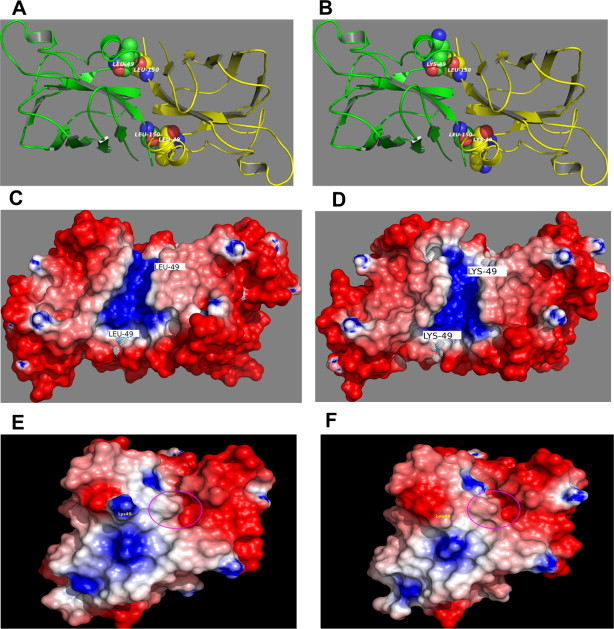
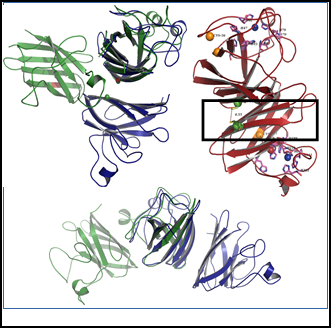
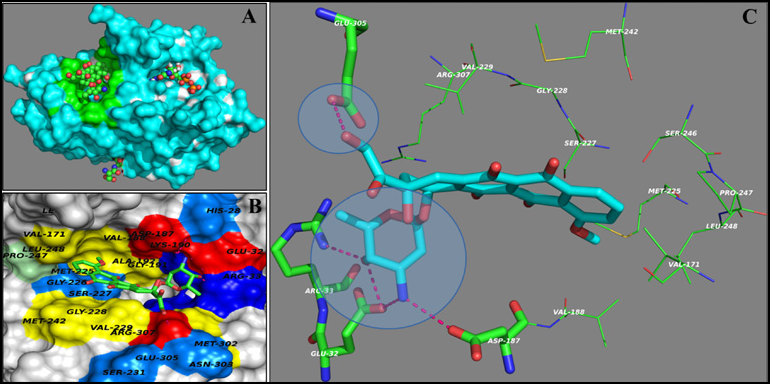
2024
- Agarwal, S., Parija, M., Naik, S., Kumari, P., Mishra, S. K., Adhya, A. K., Kashaw, S. K., Dixit, A. Dysregulated gene subnetworks in breast invasive carcinoma reveal novel tumor suppressor genes, Scientific Reports, 2024 (Just accepted)
2023
- Mohanty, S., Mishra, B.K., Dasgupta, M., Acharya, G., Singh, S., Naresh, P., Bhue, S., Dixit, A., Sarkar, A., Sahoo, M.R. Deciphering phenotyping, DNA barcoding, and RNA secondary structure predictions in eggplant wild relatives provide insights for their future breeding strategies. Scientific Reports, 13, 13829, 2023.
- Abhishek, Kondyarpu; Mohanta, Bineet Kumar; Kumari, Pratima; Dixit, Anshuman; Puppala, Venkat Ramchander; GeMemiOM—the first curated database on Genes, putative Methylation study targets, and microRNA targets for Otitis Media, Journal of Genetics and Genomics, 1673-8527(23)00162-5, 2023.
- Nagaraj, Viswanathan Arun; Ghosh, Sourav; Kundu, Rajib; Chandana, Manjunatha; Das, Rahul; Anand, Aditya; Beura, Subhashree; Bobde, Ruchir; Jain, Vishal; Prabhu, Sowmya; Behera, Prativa; Mohanty, Akshaya; Mahabala, Chakrapani; Satyamoorthy, Kapaettu; Suryawanshi, Amol; Dixit, Anshuman; Padmanaban, Govindarajan, “Distinct Evolution of Type I Glutamine Synthetase in Plasmodium and its Species-specific Requirement” Nature Communications, 14, 4216, 2023.
- Nayak, J.; Jena, Soumya Ranjan; Kumar, Sugandh; Kar, Sujata; Dixit, Anshuman; Samanta, Luna, Human sperm proteome reveals the effect of environmental borne seminal polyaromatic hydrocarbons exposome in etiology of idiopathic male factor infertility, Frontiers in Cell and Developmental Biology-Molecular and Cellular Pathology. Just Published, 2023.
2022
- Kumari, Pratima; Kumar, Sugandh; Sethy, Madhusmita; Bhue, Shyamlal; Mohanta, Bineet Kumar; Dixit, Anshuman, Identification of Therapeutically Potential Targets and their Ligands for the Treatment of OSCC, Frontiers in Oncology, 2022.
- Swain, Nirlipta; Samanta, Luna; Goswami, Chandan; Kar, Sujata; Majhi, Rakesh Kumar; Kumar, Sugandh; Dixit, Anshuman, TRPV1 channel in spermatozoa is a molecular target for ROS mediated sperm dysfunction and differentially expressed in both natural and ART pregnancy failure, Frontiers in Cell and Developmental Biology, 2022.
- Nayak, J.; Jena, SR; Kumar, S.; Kar, S.; Dixit, A.; Samanta, L. Comparative proteome profiling of seminal components reveal impaired immune cell signaling as paternal contributors in recurrent pregnancy loss patients, American Journal of Reproductive Immunology, e13613, doi: 10.1111/aji.13613.2022.
- Kumar, Anoop; Sahu, Utkarsha; Kumari, Pratima; Dixit, Anshuman; Khare, Prashant; Designing of multi-epitope chimeric vaccine using immunoinformatic platform by targeting oncogenic strain HPV 16 and 18 against cervical cancer, Scientific Reports, 12:9521, 2022.
- Kumari, Pratima; Debta, Priyanka; Dixit, Anshuman; Oral Potentially Malignant Disorders: Etiology, Pathogenesis, and Transformation Into Oral Cancer. Frontiers in Pharmacology, 13: 825266, 2022.
- Singh, Deepika; Mohapatra, Priyanka; Kumar, Sugandh; Behera, Somalisa; Dixit, Anshuman; Sahoo, Sanjeeb Kumar; Nimbolide-encapsulated PLGA nanoparticles induces Mesenchymal-to-Epithelial Transition by dual inhibition of AKT and mTOR in pancreatic cancer stem cells, Toxicology in Vitro, 79 105293 5, 2022.
- Nayak, Jasmine; Jena, Soumya Ranjan; Kumar, Sugandh; Kar, Sujata; Dixit, Anshuman; Samanta, Luna; Association of seminal polyaromatic hydrocarbons exposome with idiopathic male factor infertility: A proteomic insight into sperm function, bioRxiv, 2022.
- S Khan, R Sinha, S Sarkar, A Dixit, S Routray, Microbial Dysbiosis in Oral Cancer In Microbes and Oral Squamous Cell Carcinoma, 95-106, 2022.
- S Khan, R Sinha, S Routray, A Dixit, Implications and Future Perspectives In Microbes and Oral Squamous Cell Carcinoma, 163-172, 2022.
- S Khan, R Sinha, A Dixit, Microbial “OMICS” in Oral Cancer In Microbes and Oral Squamous Cell Carcinoma, 149-161, 2022
2021
- Agarwal, Shivangi; Sau, Samaresh; Iyer, Arun K; Dixit, Anshuman; Kashaw, Sushil K; Multiple strategies for the treatment of invasive breast carcinoma: a comprehensive prospective, Drug Discovery Today, 2021. doi:10.1016/j.drudis.2021.10.008
- Fatma, Sana; Kumar, Ravindra; Dixit, Anshuman; Swain, Rajeeb K; Expression of two uncharacterized protein coding genes in zebrafish lateral line system, International Journal of Developmental Biology, 2021.
- Dabas, Preeti; Dhingra, Yukti; Sweta, Kumari; Chakrabarty, Mohima; Singhal, Ritwik; Tyagi, Prasidhi; Behera, Pabitra Mohan; Dixit, Anshuman; Bhattacharjee, Saikat; Sharma, Nimisha; Arabidopsis thaliana possesses two novel ELL Associated Factor (EAF) Homologs, IUBMB Life, 2021. https://doi.org/10.1002/iub.2513
- Kumar, Sugandh; Singh, Bharati; Kumari, Pratima; Kumar, Preethy V; Agnihotri, Geetanjali; Khan, Shaheerah; Beuria, Tushar Kant; Syed, Gulam Hussain; Dixit, Anshuman, Identification of multipotent drugs for COVID-19 therapeutics with the evaluation of their SARS-CoV2 inhibitory activity, Computational and Structural Biotechnology Journal, 19, 1998-2017, 2021.
- Khan, Shaheerah; Jha, Atimukta; Panda, Amaresh C.; Dixit, Anshuman, Cancer-associated circRNA-miRNA-mRNA regulatory networks: A meta-analysis, Frontiers in Molecular Biosciences, 8, 337, 2021.
- Shriwas, Omprakash; Arya, Rakesh; Mohanty, Sibasish; Mohapatra, Pallavi; Kumar, Sugandh; Rath, Rachna; Kaushik, Sandeep Rai; Pahwa, Falak; Murmu, Krushna Chandra; Majumdar, Saroj Kumar Das; Muduly, Dillip Kumar; Dixit, Anshuman; Prasad, Punit; Nanda, Ranjan K.; Dash, Rupesh, RRBP1 rewires cisplatin resistance in oral squamous cell carcinoma by regulating Hippo pathway. Br J Cancer, 124, 2004–2016, 2021.
- Routray, Samapika; Kumar, Ravindra; Datta, Keshava K; Puttamallesh, Vinuth N; Chatterjee, Aditi; Gowda, Harsha; Mohanty, Neeta; Dash, Rupesh; Dixit, Anshuman, An integrated approach for identification of a panel of candidate genes arbitrated for invasion and metastasis in oral squamous cell carcinoma, Scientific Reports, 11, 1, 2021.
- Koochana, Prashanth Kumar; Mohanty, Abhinav; Parida, Akankshika; Behera, Narmada; Behera, Pabitra Mohan; Dixit, Anshuman; Behera, Rabindra K, Flavin-mediated reductive iron mobilization from frog M and Mycobacterial ferritins: impact of their size, charge and reactivities with NADH/O 2, Journal of Biological Inorganic Chemistry, 26, 265-281, 2021.
- Nayak, A.; Kumar, S.; Singh, S; Bhattacharyya, A.; Dixit, A.; Roychowdhury, A., Oncogenic Potential of ATAD2 in Stomach Cancer and Insights into the Protein-protein Interactions at Its AAA+ATPase Domain and Bromodomain. Journal of Biomolecular Structure and Dynamics, 13, 1-17, 2021.
- Jena, Soumya R; Nayak, Jasmine; Kumar, Sugandh; Kar, Sujata; Dixit, Anshuman; Samanta, Luna, Paternal contributors in recurrent pregnancy loss: Cues from comparative proteome profiling of seminal extracellular vesicles, Molecular reproduction and development, 88, 1, 96-112, 2021.
2020
- Swain, N.; Samanta, L.; Agarwal, A.; Kumar, S.; Dixit, A.; Gopalan, B.; Durairajanayagam, D.; Sharma, R.; Pushparaj, P. N.; Baskaran, S., Aberrant upregulation of compensatory redox molecular machines may contribute to sperm dysfunction in infertile men with unilateral varicocele: A proteomic insight. Antioxidants & Redox Signaling 2020, 32 (8), 504-521.
- Raghav, S.; Ghosh, A.; Turuk, J.; Kumar, S.; Jha, A.; Madhulika, S.; Priyadarshini, M.; Biswas, V. K.; Shyamli, P. S.; Singh, B., … Dixit A, Analysis of Indian SARS-CoV-2 Genomes Reveals Prevalence of D614G Mutation in Spike Protein Predicting an Increase in Interaction With TMPRSS2 and Virus Infectivity, Frontiers in Microbiology, 2020, 11, 2847.
- Mishra, M.; Agarwal, S.; Dixit, A.; Mishra, V. K.; Kashaw, V.; Agrawal, R. K.; Kashaw, S. K., Integrated computational investigation to develop molecular design of quinazoline scaffold as promising inhibitors of plasmodium lactate dehydrogenase. Journal of Molecular Structure 2020, 1207, 127808.
- Kumar, S.; Patnaik, S.; Dixit, A., Predictive models for stage and risk classification in head and neck squamous cell carcinoma (HNSCC). PeerJ 2020, 8, e9656.
- Kumar, S.; Kumari, P.; Agnihotri, G.; VijayKumar, P.; Khan, S.; Syed, G. H.; Dixit, A., Identification of Drugs Targeting Multiple Viral and Human Proteins Using Computational Analysis for Repurposing Against COVID-19. ChemRxiv, 2020.
- Das, D.; Das, A.; Sahu, M.; Mishra, S. S.; Khan, S.; Bejugam, P. R.; Rout, P. K.; Das, A.; Bano, S.; Mishra, G. P., Identification and Characterization of Circular Intronic RNAs Derived from Insulin Gene. International journal of molecular sciences 2020, 21 (12), 4302.
- Chanwala, J.; Satpati, S.; Dixit, A.; Parida, A.; Giri, M. K.; Dey, N., Genome-wide identification and expression analysis of WRKY transcription factors in pearl millet (Pennisetum glaucum) under dehydration and salinity stress. BMC genomics 2020, 21 (1), 1-16.
- Agarwal, S.; Naik, S.; Kumari, P.; Mishra, S. K.; Adhya, A. K.; Kashaw, S. K.; Dixit, A., Stagewise identification of significantly mutated gene clusters in major molecular classes of breast invasive carcinoma. bioRxiv 2020.
- Agarwal, S.; Dixit, A.; Kashaw, S. K., Ligand and structure based virtual screening of chemical databases to explore potent small molecule inhibitors against breast invasive carcinoma using recent computational technologies. Journal of Molecular Graphics and Modelling 2020, 107591.
2019
- Satapathy, S.; Behera, P. M.; Tanty, D. K.; Srivastava, S.; Thatoi, H.; Dixit, A.; Sahoo, S. L., Isolation and molecular identification of pectinase producing Aspergillus species from different soil samples of Bhubaneswar regions. bioRxiv 2019, 837112.
- Koochana, P. K.; Mohanty, A.; Subhadarshanee, B.; Satpati, S.; Naskar, R.; Dixit, A.; Behera, R. K., Phenothiazines and phenoxazines: as electron transfer mediators for ferritin iron release. Dalton transactions 2019, 48 (10), 3314-3326.
- Gupta, D.; Satpati, S.; Dixit, A.; Ranjan, R., Fabrication of biobeads expressing heavy metal-binding protein for removal of heavy metal from wastewater. Applied microbiology and biotechnology 2019, 103 (13), 5411-5420.
- Dash, U. C.; Kanhar, S.; Dixit, A.; Dandapat, J.; Sahoo, A. K., Isolation, identification, and quantification of Pentylcurcumene from Geophila repens: A new class of cholinesterase inhibitor for Alzheimer’s disease. Bioorganic chemistry 2019, 88, 102947.
2018
- Punganuru, S. R.; Madala, H. R.; Mikelis, C. M.; Dixit, A.; Arutla, V.; Srivenugopal, K. S., Conception, synthesis, and characterization of a rofecoxib-combretastatin hybrid drug with potent cyclooxygenase-2 (COX-2) inhibiting and microtubule disrupting activities in colon cancer cell culture and xenograft models. Oncotarget 2018, 9 (40), 26109.
- Koochana, P. K.; Mohanty, A.; Das, S.; Subhadarshanee, B.; Satpati, S.; Dixit, A.; Sabat, S. C.; Behera, R. K., Releasing iron from ferritin protein nanocage by reductive method: The role of electron transfer mediator. Biochimica et Biophysica Acta (BBA)-General Subjects 2018, 1862 (5), 1190-1198.
2017
- Singh, A.; Raghuwanshi, K.; Patel, V. K.; Jain, D. K.; Veerasamy, R.; Dixit, A.; Rajak, H., Assessment of 5-substituted isatin as surface recognition group: design, synthesis, and antiproliferative evaluation of hydroxamates as novel histone deacetylase inhibitors. Pharmaceutical Chemistry Journal 2017, 51 (5), 366-374.
- Satpati, S.; Manohar, K.; Acharya, N.; Dixit, A., Comparative molecular dynamics studies of heterozygous open reading frames of DNA polymerase eta (η) in pathogenic yeast Candida albicans. Scientific reports 2017, 7 (1), 1-14.
- Kumar, R.; Samal, S. K.; Routray, S.; Dash, R.; Dixit, A., Identification of oral cancer related candidate genes by integrating protein-protein interactions, gene ontology, pathway analysis and immunohistochemistry. Scientific reports 2017, 7 (1), 1-18.
- Behera, D. K.; Behera, P. M.; Acharya, L.; Dixit, A., Development and validation of pharmacophore and QSAR models for influenza PB2 inhibitors. Chemical Biology Letters 2017, 4 (1), 1-8.
- Behera, D.; Behera, P.; Acharya, L.; Dixit, A., Pharmacophore modelling, virtual screening and molecular docking studies on PLD1 inhibitors. SAR and QSAR in Environmental Research 2017, 28 (12), 991-1009.
2016
- Satpati, S.; Behera, P.; Dixit, A., IDENTIFICATION OF LipY INHIBITORS AS ANTITUBERCULAR AGENTS USING STEPWISE VIRTUAL SCREENING. International Journal of Pharmaceutical, Chemical & Biological Sciences 2016, 6 (4).
- Panda, A.; Satpati, S.; Dixit, A.; Pal, S., Novel homologated-apio adenosine derivatives as A 3 adenosine receptor agonists: design, synthesis and molecular docking studies. RSC advances 2016, 6 (14), 11233-11239.
- Mishra, P.; Satpati, S.; Baral, S. K.; Dixit, A.; Sabat, S. C., S95C substitution in CuZn-SOD of Ipomoea carnea: impact on the structure, function and stability. Molecular BioSystems 2016, 12 (10), 3017-3031.
- Jena, J.; Kumar, R.; Saifuddin, M.; Dixit, A.; Das, T., Anoxic–aerobic SBR system for nitrate, phosphate and COD removal from high-strength wastewater and diversity study of microbial communities. Biochemical Engineering Journal 2016, 105, 80-89.
2015
- Rajak, H.; Jain, D. K.; Singh, A.; Sharma, A. K.; Dixit, A., Ebola virus disease: past, present and future. Asian Pacific Journal of Tropical Biomedicine 2015, 5 (5), 337-343.
- Panda, P.; Taviti, A. C.; Satpati, S.; Kar, M. M.; Dixit, A.; Beuria, T. K., Doxorubicin inhibits E. coli division by interacting at a novel site in FtsZ. Biochemical Journal 2015, 471 (3), 335-346.
- Meher, B. R.; Dixit, A.; Bousfield, G. R.; Lushington, G. H., Glycosylation effects on FSH-FSHR interaction dynamics: a case study of different FSH glycoforms by molecular dynamics simulations. PloS one 2015, 10 (9), e0137897.
- Kumar, S.; Mamidi, P.; Kumar, A.; Basantray, I.; Bramha, U.; Dixit, A.; Maiti, P. K.; Singh, S.; Suryawanshi, A. R.; Chattopadhyay, S., Development of novel antibodies against non-structural proteins nsP1, nsP3 and nsP4 of chikungunya virus: potential use in basic research. Archives of virology 2015, 160 (11), 2749-2761.
- Jena, J.; Kumar, R.; Dixit, A.; Pandey, S.; Das, T., Evaluation of simultaneous nutrient and COD removal with polyhydroxybutyrate (PHB) accumulation using mixed microbial consortia under anoxic condition and their bioinformatics analysis. PloS one 2015, 10 (2), e0116230.
- Behera, P. M.; Behera, D. K.; Satpati, S.; Agnihotri, G.; Nayak, S.; Padhi, P.; Dixit, A., Molecular modeling and identification of novel glucokinase activators through stepwise virtual screening. Journal of Molecular Graphics and Modelling 2015, 57, 122-130.
- Archana, S.; Geesala, R.; Rao, N. B.; Satpati, S.; Puroshottam, G.; Panasa, A.; Dixit, A.; Das, A.; Srivastava, A. K., Development of constrained tamoxifen mimics and their antiproliferative properties against breast cancer cells. Bioorganic & medicinal chemistry letters 2015, 25 (3), 680-684.
2014
- Rajak, H.; Singh, A.; Raghuwanshi, K.; Kumar, R.; Dewangan, P.; Veerasamy, R.; Sharma, P.; Dixit, A.; Mishra, P., A structural insight into hydroxamic acid based histone deacetylase inhibitors for the presence of anticancer activity. Current medicinal chemistry 2014, 21 (23), 2642-2664.
- Mishra, P.; Dixit, A.; Ray, M.; Sabat, S. C., Mechanistic study of CuZn-SOD from Ipomoea carnea mutated at dimer interface: Enhancement of peroxidase activity upon monomerization. Biochimie 2014, 97, 181-193.
- Dixit, A.; Verkhivker, G. M., Structure-functional prediction and analysis of cancer mutation effects in protein kinases. Computational and mathematical methods in medicine 2014, 2014.
- D Sengupta, D. B., A Dixit, BS Sahoo, S Biswas, G Biswas, SK Mishra, ERRβ Signaling Involves FST and BCAS2 for Inhibiting Cellular Proliferation in Breast Cancer Cells. British journal of cancer 2014, 110 (8), 2144-2158.
- Beesu, M.; Malladi, S. S.; Fox, L. M.; Jones, C. D.; Dixit, A.; David, S. A., Human Toll-like receptor 8-selective agonistic activities in 1-alkyl-1 H-benzimidazol-2-amines. Journal of medicinal chemistry 2014, 57 (17), 7325-7341.
2013
- Rajak, H.; Kumar Dewangan, P.; Patel, V.; Kumar Jain, D.; Singh, A.; Veerasamy, R.; Chander Sharma, P.; Dixit, A., Design of combretastatin A-4 analogs as tubulin targeted vascular disrupting agent with special emphasis on their cis-restricted isomers. Current pharmaceutical design 2013, 19 (10), 1923-1955.
- Kokatla, H. P.; Sil, D.; Malladi, S. S.; Balakrishna, R.; Hermanson, A. R.; Fox, L. M.; Wang, X.; Dixit, A.; David, S. A., Exquisite selectivity for human toll-like receptor 8 in substituted furo [2, 3-c] quinolines. Journal of medicinal chemistry 2013, 56 (17), 6871-6885.
- Behera, P. M.; Behera, D. K.; Panda, A.; Dixit, A.; Padhi, P., In Silico Expressed Sequence Tag Analysis in Identification of Probable Diabetic Genes as Virtual Therapeutic Targets. BioMed Research International 2013, 2013.
2012
- Dixit, A.; Verkhivker, G. M., Integrating ligand-based and protein-centric virtual screening of kinase inhibitors using ensembles of multiple protein kinase genes and conformations. Journal of chemical information and modeling 2012, 52 (10), 2501-2515.
- Dixit, A.; Verkhivker, G. M., Probing molecular mechanisms of the Hsp90 chaperone: Biophysical modeling identifies key regulators of functional dynamics. PloS one 2012, 7 (5), e37605.
- Dixit, A.; Verkhivker, G., Probing Molecular Mechanisms of the Hsp90 Chaperone: Biophysical Modeling Identifies Key Regulators of Functional. 2012.
- Chand, S.; Mehta, N.; Singh Bahia, M.; Dixit, A.; Silakari, O., Protein kinase C-theta inhibitors: a novel therapy for inflammatory disorders. Current pharmaceutical design 2012, 18 (30), 4725-4746.
- Behera, D. K.; Behera, P. M.; Acharya, L.; Dixit, A.; Padhi, P., In silico biology of H1N1: molecular modelling of novel receptors and docking studies of inhibitors to reveal new insight in flu treatment. Journal of Biomedicine and Biotechnology 2012, 2012.
2011
- Matts, R. L.; Dixit, A.; Peterson, L. B.; Sun, L.; Voruganti, S.; Kalyanaraman, P.; Hartson, S. D.; Verkhivker, G. M.; Blagg, B. S., Elucidation of the Hsp90 C-terminal inhibitor binding site. ACS chemical biology 2011, 6 (8), 800-807.
- Matts, R. L.; Brandt, G. E.; Lu, Y.; Dixit, A.; Mollapour, M.; Wang, S.; Donnelly, A. C.; Neckers, L.; Verkhivker, G.; Blagg, B. S., A systematic protocol for the characterization of Hsp90 modulators. Bioorganic & medicinal chemistry 2011, 19 (1), 684-692.
- Matts, R.; Dixit, A.; Peterson, L.; Sun, L.; Voruganti, S.; Kalyanaraman, P.; Hartson, S.; Verkhivker, G.; Blagg, B., Elucidation of the Hsp90 C-terminal inhibitor binding site, ACS Chem. , Article ASAP, doi 2011, 10.
- Hübner, M.; Dixit, A.; Mou, T.-C.; Lushington, G. H.; Pinto, C.; Gille, A.; Geduhn, J.; König, B.; Sprang, S. R.; Seifert, R., Structural basis for the high-affinity inhibition of mammalian membranous adenylyl cyclase by 2′, 3′-o-(N-methylanthraniloyl)-inosine 5′-triphosphate. Molecular pharmacology 2011, 80 (1), 87-96.
- Dixit, A.; Verkhivker, G. M., The energy landscape analysis of cancer mutations in protein kinases. PloS one 2011, 6 (10), e26071.
- Dixit, A.; Verkhivker, G. M., Computational modeling of allosteric communication reveals organizing principles of mutation-induced signaling in ABL and EGFR kinases. PLoS computational biology 2011, 7 (10), e1002179.
Older:
- Verkhivker, G. M.; Dixit, A.; Morra, G.; Colombo, G., Structural and computational biology of the molecular chaperone Hsp90: from understanding molecular mechanisms to computer-based inhibitor design. Current topics in medicinal chemistry 2009, 9 (15), 1369-1385.
- Dixit, A.; Yi, L.; Gowthaman, R.; Torkamani, A.; Schork, N. J.; Verkhivker, G. M., Sequence and structure signatures of cancer mutation hotspots in protein kinases. PloS one 2009, 4 (10), e7485.
- Dixit, A.; Verkhivker, G. M., Hierarchical modeling of activation mechanisms in the ABL and EGFR kinase domains: thermodynamic and mechanistic catalysts of kinase activation by cancer mutations. PLoS Comput Biol 2009, 5 (8), e1000487.
- Dixit, A.; Torkamani, A.; Schork, N. J.; Verkhivker, G., Computational modeling of structurally conserved cancer mutations in the RET and MET kinases: the impact on protein structure, dynamics, and stability. Biophysical Journal 2009, 96 (3), 858-874.
- Saxena, A.; Alam, I.; Dixit, A.; Saxena, M., Internet resources in GPCR modelling. SAR and QSAR in Environmental Research 2008, 19 (1-2), 11-25.
- Roy, K. K.; Dixit, A.; Saxena, A. K., An investigation of structurally diverse carbamates for acetylcholinesterase (AChE) inhibition using 3D-QSAR analysis. Journal of Molecular Graphics and Modelling 2008, 27 (2), 197-208.
- Misra, P.; Khaliq, T.; Dixit, A.; SenGupta, S.; Samant, M.; Kumari, S.; Kumar, A.; Kushawaha, P. K.; Majumder, H.; Saxena, A. K., Antileishmanial activity mediated by apoptosis and structure-based target study of peganine hydrochloride dihydrate: an approach for rational drug design. Journal of antimicrobial chemotherapy 2008, 62 (5), 998-1002.
- Geronikaki, A.; Eleftheriou, P.; Vicini, P.; Alam, I.; Dixit, A.; Saxena, A., 2-Thiazolylimino/heteroarylimino-5-arylidene-4-thiazolidinones as new agents with SHP-2 inhibitory action. Journal of medicinal chemistry 2008, 51 (17), 5221-5228.
- Dixit, A.; Saxena, A. K., QSAR analysis of PPAR-γ agonists as anti-diabetic agents. European journal of medicinal chemistry 2008, 43 (1), 73-80.
- Team, N.-B., BioSuite: A comprehensive bioinformatics software package (A unique industry—academia collaboration). Current Science 2007, 29-38.
- Prathipati, P.; Dixit, A.; Saxena, A. K., Computer-aided drug design: integration of structure-based and ligand-based approaches in drug design. Current Computer-Aided Drug Design 2007, 3 (2), 133-148.
- Lakshmi, M.; Dixit, A.; Saxena, A. K., QSAR studies on cis-hexa and tetra-hydrophthalazinones: A new class of selective PDE-4 inhibitors. 2006.
- Rathi, L.; Kashaw, S. K.; Dixit, A.; Pandey, G.; Saxena, A. K., Pharmacophore identification and 3D-QSAR studies in N-(2-benzoyl phenyl)-L-tyrosines as PPARγ agonists. Bioorganic & medicinal chemistry 2004, 12 (1), 63-69.
- Dixit, A.; Kashaw, S. K.; Gaur, S.; Saxena, A. K., Development of CoMFA, advance CoMFA and CoMSIA models in pyrroloquinazolines as thrombin receptor antagonist. Bioorganic & medicinal chemistry 2004, 12 (13), 3591-3598.
- Silakari, O.; Dixit, A.; Kohli, D.; Chaturvedi, S., QSAR analysis of 4, 5-diarylpyrroles with cyclooxygenase-2 inhibitory activity. Indian journal of pharmaceutical sciences 2001, 63 (6), 518-521.
Book chapter
- Behera, Pabitra Mohan; Dixit, Anshuman; The story of kinase inhibitors development with special reference to allosteric site, Drug Design: Principles and Applications, 57-68, 2017, Springer, Singapore.
International Patent
Gaur, Stuti; Fatima, Zeeshan; Dixit, Anshuman; Ali, Zahid; Surin, William Rascan; Kappor, Kapil; Bhutani, Kanta; Ansari, Mohammed Salim; Dikshit, Madhu; Saxena, Anil Kumar; , 2-Alkyl/aryl sulphonyl-1, 2, 3, 4-tetrahydro-9h-pyrido (3, 4-b) indole-3-carboxylic acid esters/amides useful as antithrombotic agents, 2009, US Patent 7,601,838.
|