Updated publication list can be found in GOOGLE SCHOLAR and PUBMED
* Corresponding Author Publications; # Equal Contribution
ILS Publications
- Das A; Rout PK; Gorospe M; Panda AC*. Rolling Circle cDNA Synthesis Uncovers Circular RNA Splice variants. Int. J. Mol. Sci. 2019, 20(16), 3988
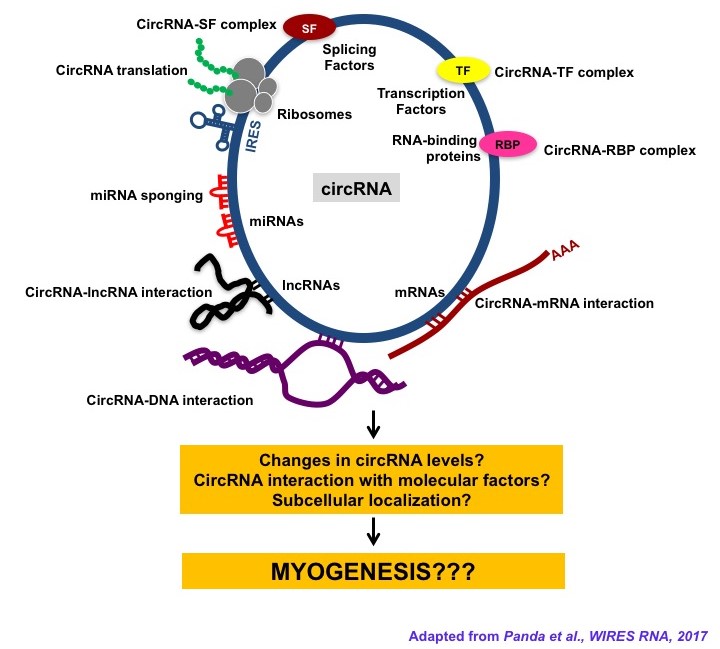
- Das A; Das A; Das D; Abdelmohsen K; Panda AC*. Circular RNAs in myogenesis. BBA-Gene Regulatory Mechanisms. 2019; DOI: 10.1016/j.bbagrm.2019.02.011.
- Munk R; Martindale JL; Yang X; Yang JH; Grammatikakis I; Di Germanio C; Mitchell SJ; de Cabo R; Lehrmann E; Zhang Y; Becker KG; Raz V; Gorospe M*; Abdelmohsen K; Panda AC*. Loss of miR-451a enhances SPARC production during myogenesis. PLoS One. 2019; 14(3):e0214301.
- Pandey P; Rout PK; Das A; Gorospe M*; Panda AC*. RPAD (RNase R Treatment, Polyadenylation, and Poly(A)+ RNA Depletion) Method to Isolate Highly Pure Circular RNA. 2019 Feb 15;155:41-48.
- Das D; Das A; Panda AC*. Emerging role of long noncoding RNAs and circular RNAs in pancreatic β cells. Non-coding RNA Investigation. 2018;2:69.
- Panda AC*. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018; 1087:67-79.
- Das A, Gorospe M, Panda AC*. The coding potential of circRNAs. Aging (Albany NY). 2018 Sep 13; 10(9):2228-2229.
- Panda AC* and Gorospe M. Identifying intronic circRNAs: progress and challenges. Non-coding RNA Investig 2018;2:34.
- Panda AC* and Gorospe M. Detection and Analysis of Circular RNAs by RT-PCR. Bio-protocol. 2018 Mar 20; 8(6): e2775.
Previous Publications
-
Panda AC, Dudekula DB, Abdelmohsen K, Gorospe M. Analysis of Circular RNAs Using the Web Tool CircInteractome. Methods in Molecular Biology, vol 1724; 2018 Jan 11.
-
Panda AC, Abdelmohsen K, and Gorospe M. SASP Regulation by Noncoding RNA. Mechanism of Aging and Development. 2017 May 11
-
Munk R, Panda AC, Grammatikakis I, Gorospe M and Abdelmohsen K. Senescence-associated miRNAs. International Review of Cell and Molecular Biology. 2017 April 28.
-
Panda AC*#, De S#, Grammatikakis I, Munk R, Yang X, Piao Y. Dudekula DB, Abdelmohsen K*, and Gorospe M. High-purity circular RNA isolation method (RPAD) reveals vast collection of intronic circRNAs (IcircRNAs). Nucleic Acids Res. 2017 Jul 7; 45(12): e116. #Equal Contribution
-
Panda AC*#, Grammatikakis I*#, Kim KM, De S, Martindale JL, Munk R, Yang X, Abdelmohsen K, and Gorospe M. Identification of Senescence-Associated Circular RNAs (SAC-RNAs) reveals senescence suppressor CircPVT1. Nucleic Acids Res. 2017 Apr 20;45(7):4021-4035. #Equal Contribution
-
Panda AC, Abdelmohsen K, Gorospe M. RT-qPCR detection of senescence-associated circular RNAs. Methods in Molecular Biology. 2017; 1534:79-87.
-
Panda AC, Grammatikakis I, Munk R, Gorospe M, Abdelmohsen K. Emerging roles and context of circular RNAs. Wiley Interdiscip Rev RNA. 2017 Mar: 8(2).
-
Abdelmohsen K#, Panda AC#, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, and Gorospe M. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biology. 2017 Mar 4;14(3):361-369.
-
Panda AC*, Martindale JL, Gorospe M. Polysome Fractionation to Analyze mRNA Distribution Profiles. Bio-Protocol. 2017 Feb 5, Vol 7, Iss 03.
-
Panda AC*, Martindale JL, Gorospe M. Affinity Pulldown of Biotinylated RNA for Detection of Protein-RNA Complexes. Bio-Protocol. 2016 Dec 20, Vol 6, Iss 24.
-
Di Francesco A, Di Germanio C, Panda AC, Huynh P, Peaden R, Navas-Enamorado I, Bastian P, Lehrmann E, Diaz-Ruiz A, Ross D, Siegel D, Martindale JL, Bernier M, Gorospe M, Abdelmohsen K, de Cabo R. Novel RNA-binding activity of NQO1 promotes SERPINA1 mRNA translation. Free Radic Biol Med. 2016 Aug 8;99:225-233.
-
Noh JH, Kim KM, Abdelmohsen K, Yoon JH, Panda AC, Ghosh P, Munk R, Curtis J, Moad CA, Indig FE, Paula WD, Dudekula DB, De S, Yang X, Martindale JL, de Cabo R, and Gorospe M. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016 May 15;30(10):1224-39.
-
Grammatikakis I, Peisu Z, Panda AC, Kim J, Maudsley S, Abdelmohsen K, Yang X, Martindale JL, Motiño O, Hutchison ER, Mattson MP, and Gorospe M. Alternative splicing of neuronal differentiation factor TRF2 regulated by HNRNPH1/H2. Cell Reports. 2016 May 3;15(5):926-934.
-
Panda AC*, Abdelmohsen K, Martindale JL, Di Germanio C, Yang X, Grammatikakis I, Noh JH, Zhang Y, Lehrmann E, Dudekula DB, De S, Becker KG, White EJ, Wilson GM, de Cabo R, Gorospe M. Novel RNA-binding activity of MYF5 enhances Ccnd1/Cyclin D1 mRNA translation during myogenesis. Nucleic Acids Res. 2016 Mar 18;44(5):2393-408.
-
Dudekula DB#, Panda AC#, Grammatikakis I, De S, Abdelmohsen K, and Gorospe M. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biology, 2016, Jan 2;13(1):34-42.
-
Abdelmohsen K#, Panda AC#, De S#, Grammatikakis I, Kim J, Ding J, Noh JH, Kim KM, Mattison JA, de Cabo R, Gorospe M. Circular RNAs in monkey muscle: age-dependent changes. Aging (Albany NY). 2015 Nov; 7(11): 903-910.
-
Lee KP, Shin YJ, Panda AC, Abdelmohsen K, Kim JY, Lee SM, Bahn YJ, Choi JY, Kwon ES, Baek SJ, Kim SY, Gorospe M, Kwon KS. miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4. Genes Dev. 2015 Aug 1;29(15):1605-17.
-
Grammatikakis I#, Panda AC#, Abdelmohsen K, Gorospe M. Long noncoding RNAs (lncRNAs) and the molecular hallmarks of aging. Aging (Albany NY). 2014 Dec ;6(12) :992-1009.
-
Abdelmohsen K, Panda AC, Kang MJ, Guo R, Kim J, Grammatikakis I, Yoon JH, Dudekula DB, Noh JH, Yang X, Martindale JL, Gorospe M. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014 Sep;42(15):10099-111.
-
Panda AC, Sahu I, Kulkarni SD, Martindale JL, Abdelmohsen K, Vindu A, Joseph J, Gorospe M, Seshadri V. miR-196b-mediated translation regulation of mouse insulin2 via the 5’UTR. PLoS One. 2014 Jul 8;9(7):e101084.
-
Panda AC, Abdelmohsen K, Yoon JH, Martindale JL, Yang X, Curtis J, Mercken EM, Chenette DM, Zhang Y, Schneider RJ, Becker KG, de Cabo R, Gorospe M. RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels. Mol Cell Biol. 2014 Aug;34(16):3106-19.
-
Abdelmohsen K, Panda A, Kang MJ, Xu J, Selimyan R, Yoon JH, Martindale JL, De S, Wood WH 3rd, Becker KG, Gorospe M. Senescence-associated lncRNAs: senescence-associated long noncoding RNAs. Aging Cell. 2013 Oct;12(5):890-900.
-
Panda AC, Grammatikakis I, Yoon JH, Abdelmohsen K. Posttranscriptional regulation of insulin family ligands and receptors. Int J Mol Sci. 2013 Sep 18;14(9):19202-29.
-
Chatterjee S#, Panda AC#, Berwal SK, Sreejith RK, Ritvika C, Seshadri V, Pal JK. Vimentin is a component of a complex that binds to the 5′-UTR of human heme-regulated eIF2α kinase mRNA and regulates its translation. FEBS Lett. 2013 Mar 1;587(5):474-80.
-
Kulkarni SD, Muralidharan B, Panda AC, Bakthavachalu B, Vindu A, Seshadri V. Glucose-stimulated translation regulation of insulin by the 5′ UTR-binding proteins. J Biol Chem. 2011 Apr 22;286(16):14146-56.
-
Panda AC, Kulkarni SD, Muralidharan B, Bakthavachalu B, Seshadri V. Novel splice variant of mouse insulin2 mRNA: implications for insulin expression. FEBS Lett. 2010 Mar 19;584(6):1169-73.
Other publications
-
Panda AC and Seshadri V. Mus musculus insulin2 precursor (Ins2) mRNA, partial cds, alternatively spliced. Gene Bank # GQ915612.1.
|


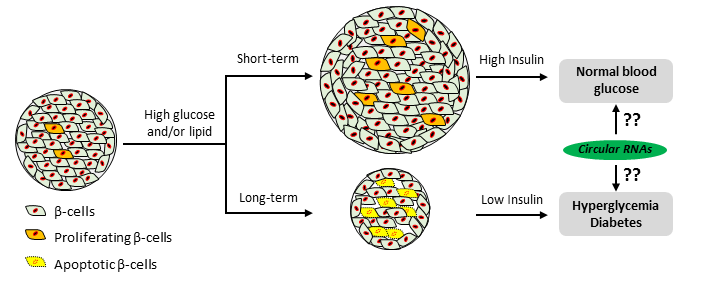
 RNA-RNA interactions in pancreatic β-cell physiology
RNA-RNA interactions in pancreatic β-cell physiology